
He has developed algorithms for protein identification since 1996, when he began graduate school in the Laboratory of John Yates. Is an associate professor of Biomedical Informatics at Vanderbilt University. The content is designed to be accessible to computer scientists and bioinformaticists who have not previously worked with proteomics data sets. This workshop introduces major elements of the protein identification and quantitation pipelines and describes a strategy for proteogenomic experiments with both RNA-Seq and proteomic data. Identifying proteins and post-translational modifications (PTMs) from tandem mass spectrometry data depends heavily on an ecosystem of algorithms that has emerged during the last decade. Evaluating impact on proteomic identification.Customizing protein sequence databases from RNA-Seq data.Proteogenomics through RNA-Seq / proteomic combination Building upon prior identifications through spectral library search.Publishing proteomes through ProteomeXchange and LabCAS.Proteomic repositories and spectral libraries Monitoring batch effects and proteomics quality control.Incorporating MS intensity in protein quantitation.
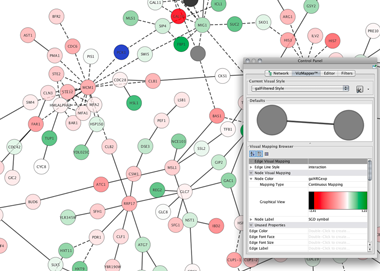
Differentiating cohorts by protein spectral counts.


 0 kommentar(er)
0 kommentar(er)
